Project information
- Keywords: Biomarkers, RNA, Docking, QM/MM, MD
- Project date: Ongoing
- Preprint URL: TBD
- Journal: TBD
Project Details
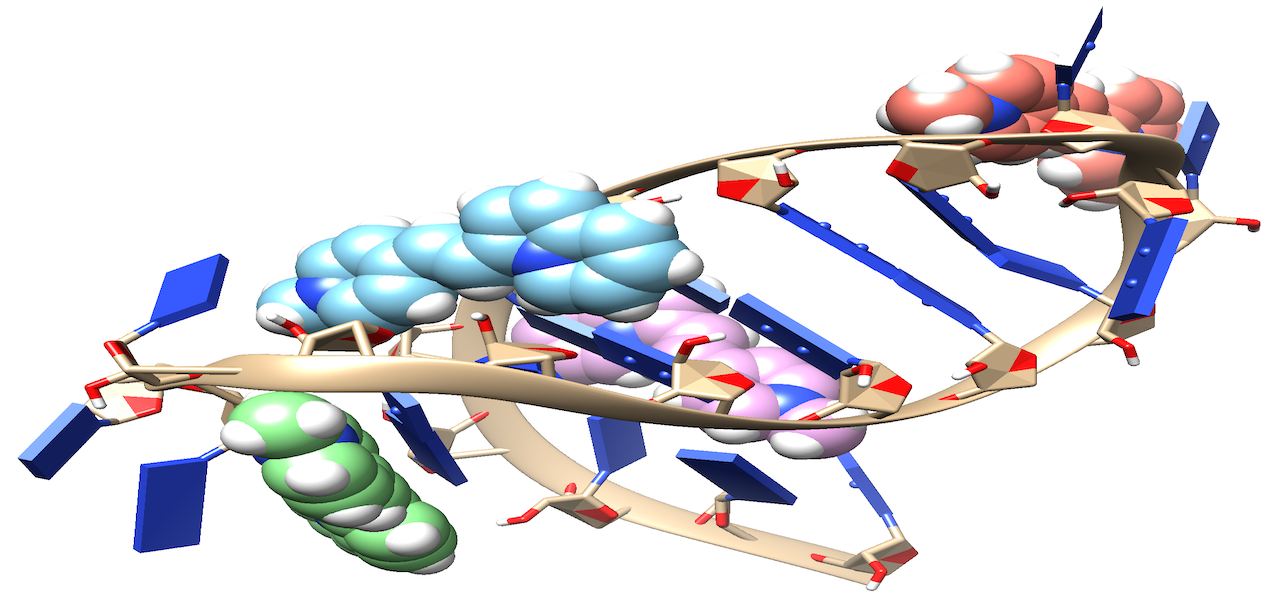
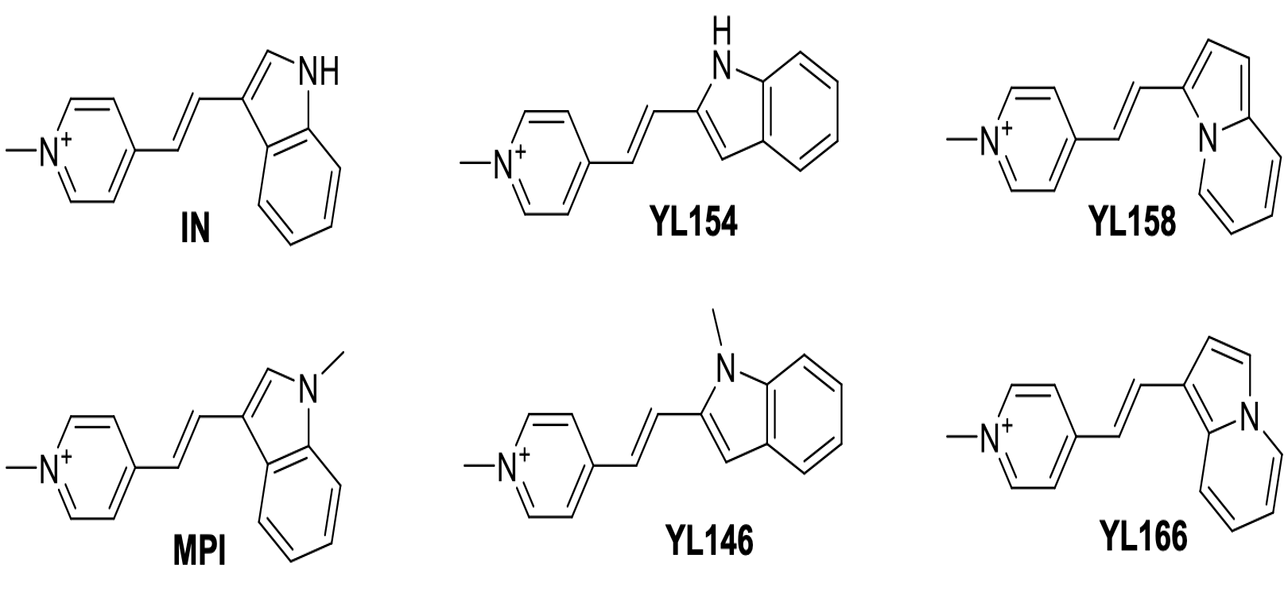
Visualizing RNA localization in living cells is crucial for understanding the corresponding protein localization. Methods for selectively visualizing RNA include fluorescence in situ hybridization (FISH), which employs fluorophore-linked RNA probes binding to RNA targets. Fluorescence imaging reveals the localized regions by measuring fluorophore emission. Ideal chromophores for in vivo experiments should have compatible absorption wavelengths, high quantum yield, bright emission, and resistance to photobleaching. Electronic structure calculations were performed on styryl-based molecules as potential RNA biomarkers. Calculations included electronic transitions, analysis of the lowest bright excited state, and characterization of electronic transitions through permanent dipole moment and natural transition orbitals (NTOs). Excited-state calculations were conducted using DFT/ωB97M-V/aug-cc-pVTZ and EOM-CCSD/aug-cc-pVDZ levels in implicit solvent (PCM) for DMSO. The Hückel method correlated biomarker structure with spectral properties. Preliminary results on chromophore interactions with RNA molecules were obtained from docking, MD simulations, and QM/MM calculations.